Proteins are the largest and most varied class of biological molecules, and they show the greatest variety of structures. Many have intricate three-dimensional folding patterns that result in a compact form, but others do not fold up at all (“natively unstructured proteins”) and exist in random conformations. The function of proteins depends on their structure, and defining the structure of individual proteins is a large part of modern Biochemistry and Molecular Biology.
To understand how proteins fold, we will start with the basics of structure, and progress through to structures of increasing complexity.
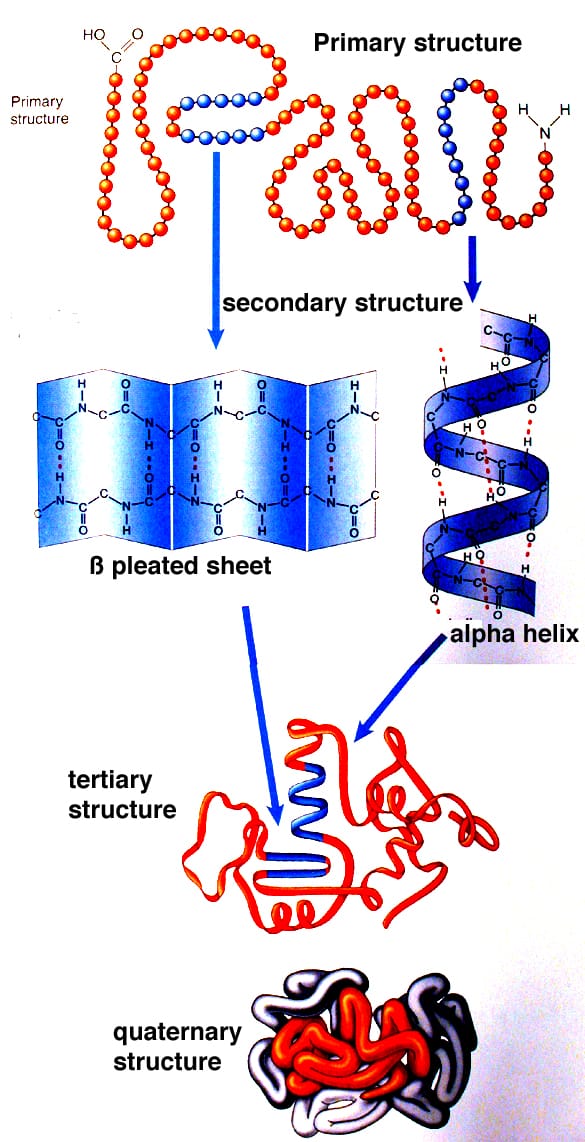
Peptide Bonds
To make a protein, amino acids are connected together by a type of amide bond called a “peptide bond”. This bond is formed between the alpha amino group of one amino acid and the carboxyl group of another in a condensation reaction. When two amino acids join, the result is called a dipeptide, three gives a tripeptide, etc. Multiple amino acids result in a polypeptide (often shortened to “peptide”).
Because water is lost in the course of creating the peptide bond, individual amino acids are referred to as “amino acid residues” once they are incorporated. Another property of peptides is polarity: the two ends are different. One end has a free amino group (called the “N-terminal”) and the other has a free carboxyl group (“C-terminal”).
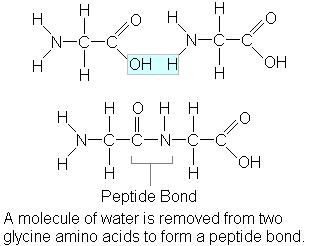
In the natural course of making a protein, polypeptides are elongated by the addition of amino acids to the C-terminal end of the growing chain. Conventionally, peptides are written N-terminal first; therefore gly-ser is not the same as ser-gly or GS is not the same as SG. The connection gives rise to a repeating pattern of “NCC-NCC-NCC…” atoms along the length of the molecule. This is referred to as the “backbone” of the peptide. If stretched out, the side chains of the individual residues project outwards from this backbone.
The peptide bond is written as a single bond, but it actually has some characteristics of a double bond because of the resonance between the C-O and C-N bonds:
This means that the six atoms involved are coplanar, and that there is no free rotation around the C–N axis. This constrains the flexibility of the chain and prevents some folding patterns.
Primary Structure of Proteins
It is convenient to discuss protein structure in terms of four levels (primary to quaternary) of increasing complexity. Primary structure is simply the sequence of residues making up the protein. Thus primary structure involves only the covalent bonds linking residues together.
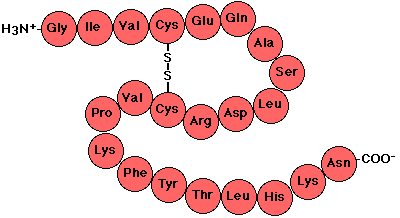
The minimum size of a protein is defined as about 50 residues; smaller chains are referred to simply as peptides. So the primary structure of a small protein would consist of a sequence of 50 or so residues. Even such small proteins contain hundreds of atoms and have molecular weights of over 5000 Daltons (Da). There is no theoretical maximum size, but the largest protein so far discovered has about 30,000 residues. Since the average molecular weight of a residue is about 110 Da, that single-chain has a molecular weight of over 3 million Daltons.
Secondary Structure
This level of structure describes the local folding pattern of the polypeptide backbone and is stabilized by hydrogen bonds between N-H and C=O groups. Various types of secondary structures have been discovered, but by far the most common is the orderly repeating forms known as the a-helix and the b sheet.
An a helix, as the name implies, is a helical arrangement of a single polypeptide chain, like a coiled spring. In this conformation, the carbonyl and N-H groups are oriented parallel to the axis. Each carbonyl is linked by a hydrogen bond to the N-H of a residue located 4 residues further on in the sequence within the same chain. All C=O and N-H groups are involved in hydrogen bonds, making a fairly rigid cylinder.
The alpha helix has precise dimensions: 3.6 residues per turn, 0.54 nm per turn. The side chains project outward and contact any solvent, producing a structure something like a bottle brush or a round hair brush. An example of a protein with many a-helical structures is the keratin that makes up human hair.
The structure of a b sheet is very different from the structure of an a-helix. In a b sheet, the polypeptide chain folds back on itself so that polypeptide strands like side by side, and are held together by hydrogen bonds, forming a very rigid structure. Again, the polypeptide N-H and C=O groups form hydrogen bonds to stabilize the structure, but unlike the a-helix, these bonds are formed between neighbouring polypeptide (b) strands.
Generally, the primary structure folds back on itself in either a parallel or antiparallel arrangement, producing a parallel or antiparallel b sheet. In this arrangement, side chains project alternately upward and downward from the sheet. The major constituent of silk (silk fibroin) consists mainly of layers of b sheet stacked on top of each another.
Other types of secondary structure. While the a helix and b sheet are by far the most common types of structure, many others are possible. These include various loops, helices and irregular conformations. A single polypeptide chain may have different regions that take on different secondary structures. In fact, many proteins have a mixture of a helices, b sheets, and other types of folding patterns to form various overall shapes.
What determines whether a particular part of a sequence will fold into one or the other of these structures? A major determinant is the interactions between side chains of the residues in the polypeptide. Several factors come into play: steric hindrance between nearby large side chains, charge repulsion between nearby similarly-charged side chains, and the presence of proline.
Proline contains a ring that constrains bond angles so that it will not fit exactly into an a-helix or b sheet. Further, there is no H on one peptide bond when proline is present, so a hydrogen bond cannot form. Another major factor is the presence of other chemical groups that interact with each other. This contributes to the next level of protein structure, the tertiary structure.
Tertiary Structure
This level of structure describes how regions of secondary structure fold together – that is, the 3D arrangement of a polypeptide chain, including a helices, b sheets, and any other loops and folds. Tertiary structure results from interactions between side chains, or between side chains and the polypeptide backbone, which are often distant in sequence. Every protein has a particular pattern of folding and these can be quite complex.
Whereas secondary structure is stabilized by H-bonding, all four “weak” forces contribute to tertiary structure. Usually, the most important force is hydrophobic interaction (or hydrophobic bonds). Polypeptide chains generally contain both hydrophobic and hydrophilic residues. Much like detergent micelles, proteins are most stable when their hydrophobic parts are buried, while hydrophilic parts are on the surface, exposed to water. Thus, more hydrophobic residues such as trp are often surrounded by other parts of the protein, excluding water, while charged residues such as asp are more often on the surface.
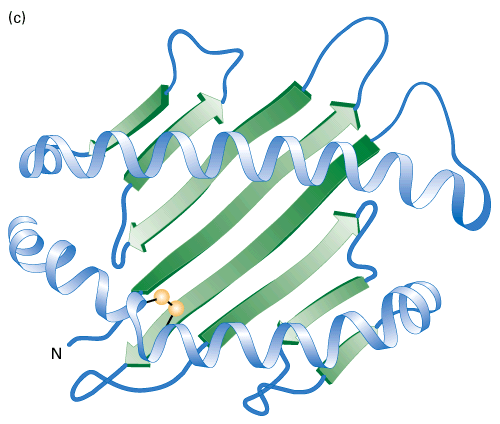
Other forces that contribute to tertiary structure are ionic bonds between side chains, hydrogen bonds, and van der Waals forces. These bonds are far weaker than covalent bonds, and it takes multiple interactions to stabilize a structure.
There is one covalent bond that is also involved in tertiary structure, and that is the disulfide bond that can form between cysteine residues. This bond is important only in non-cytoplasmic proteins since there are enzyme systems present in the cytoplasm to remove disulfide bonds.
Visualization of protein structures Because the 3D structures of proteins involve thousands of atoms in complex arrangements, various ways of depicting them so they are understood visually have been developed, each emphasizing a different property of the protein. Software tools have been written to depict proteins in many different ways, and have become essential to understanding protein structure and function.
Structural Domains of Proteins
Protein structure can also be described by a level of organization that is distinct from the ones we have just discussed. This organizational unit is the protein “domain,” and the concept of domains is extremely important for understanding tertiary structure. A domain is a distinct region (sequence of amino acids) of a protein, while a structural domain is an independently-folded part of a protein that folds into a stable structure. A protein may have many domains or consist only of a single domain. Larger proteins generally consist of connected structural domains. Domains are often separated by a loosely folded region and may create clefts between them..
Quaternary Structure
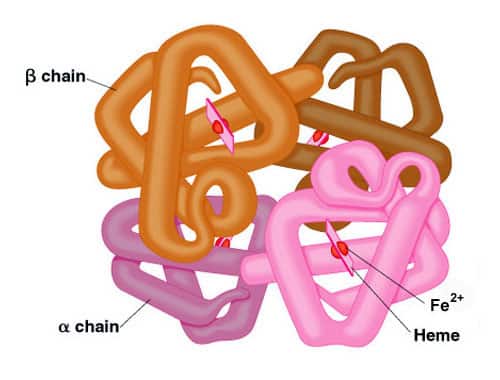
Some proteins are composed of more than one polypeptide chain. In such proteins, quaternary structure refers to the number and arrangement of the individual polypeptide chains. Each polypeptide is referred to as a subunit of the protein. The same forces and bonds that create tertiary structure also hold subunits together in a stable complex to form the complete protein.
Individual chains may be identical, somewhat similar, or totally different. As examples, CAP protein is a dimer with two identical subunits, whereas hemoglobin is a tetramer containing two pairs of non-identical (but similar) subunits. It has 2 a subunits and 2 b subunits. Secreted proteins often have subunits that are held together by disulfide bonds. Examples include tetrameric antibody molecules that commonly have two larger subunits and two smaller subunits (“heavy chains” and “light chains”) connected by disulfide bonds and noncovalent forces.
In some proteins, intertwined a-helices hold subunits together; these are called coiled-coils. This structure is stabilized by a hydrophobic surface on each a-helix that is created by a heptameric repeat pattern of hydrophilic/hydrophobic residues. The sequence of the protein can be represented as “abcdefgabcdefgabcdefg…” with positions “a” and “d” filled with hydrophobic residues such as A, V, L etc. Each a-helix has a hydrophobic surface that therefore matches the other. When the two helices coil around each other, those surfaces come together, burying the hydrophobic side chains and forming a stable structure. An example of such a protein is myosin, the motor protein found in muscle that allows contraction.
Protein Folding
How and why do proteins naturally form secondary, tertiary, and quaternary structures? This question is a very active area of research and is certainly not completely understood. A folded, biologically-active protein is considered to be in its “native” state, which is generally thought to be the conformation with the least free energy.
Proteins can be unfolded or “denatured” by treatment with solvents that disrupt weak bonds. Thus organic solvents that disrupt hydrophobic interactions, high concentrations of urea or guanidine that interfere with H-bonding, extreme pH or even high temperatures, will all cause proteins to unfold. Denatured proteins have a random, flexible conformation and usually lack biological activity. Because of exposed hydrophobic groups, they often aggregate and precipitate. This is what happens when you fry an egg.
If the denaturing condition is removed, some proteins will re-fold and regain activity. This process is called “renaturation.” Therefore, all the information necessary for folding is present in the primary structure (sequence) of the protein. During renaturation, the polypeptide chain is thought to fold up into a loose globule by hydrophobic effects, after which small regions of secondary structure form into especially favorable sequences. These sequences then interact with each other to stabilize intermediate structures before the final conformation is attained.
Many proteins have great difficulty renaturing, and proteins that assist other proteins to fold are called “molecular chaperones.” They are thought to act by reversibly masking exposed hydrophobic regions to prevent aggregation during the multi-step folding process. Proteins that must cross membranes (eg. mitochondrial proteins) must stay unfolded until they reach their destination, and molecular chaperones may protect and assist during this process.
Protein families/Types of proteins
Proteins are classified in a number of ways, according to structure, function, location and/or properties. For example, many proteins combine tightly with other substances such as carbohydrates (“glycoproteins”), lipids (“lipoproteins”), or metal ions (“metalloproteins”). The diversity of proteins that form from the 20 amino acids is greatly increased by associations such as these. Proteins that are tightly bound to membranes are called “membrane proteins”. Proteins with similar activities are given functional classifications. For example, proteins that break down other proteins are called proteases.
Because almost all proteins arise by an evolutionary process, ie. new ones are derived from old ones, they can be classified into families by their relatedness. Proteins that derive from the same ancestor are called “homologous proteins”.
Studying the sequences of homologous proteins can give clues to the structure and function of the protein. Residues that are critical for function do not change on an evolutionary timescale; they are referred to as “conserved residues”. Identifying such residues by comparing amino acid sequences often helps clarify what a protein is doing or how it is folded.
For example, the proteases trypsin and chymotrypsin are members of the “serine protease” family; so-named because of a conserved serine residue that is essential to catalyze the reaction. Trypsin and chymotrypsin contain very similar folding patterns and reaction mechanisms. Recognizing a pattern of conserved residues in protein sequences often allows scientists to deduce the function of a protein.

How do we determine which sequence of amino acids are contained within the building block of proteins?
thanks,well elaborated
Thanks, it was helpful and this was the simplest I could go to explain in my assignment.
Thanks again. 😉
@Cathy
Very interesting question. I believe in the simplest answer is the level which gives the protein specificity AND stability.
Proteins have a variety of sizes, post-translation modifications, functional groups and motif interactions. Therefore its native conformation will be controlled by those factors. A protein will fold into a 3-D structure of the lowest possible energy in vivo; the primary and secondary levels will be higher energy and less favorable. Therefore I would answer “tertiary” structure is the minimum level that proteins will require to be stable enough to be biologically active. If the protein forms a complex (which most do), then quaternary structure will be required.
Hope this helps!
I enjoyed reading your Protein Structure, which I have read 4 others and still really understand. But after reading yours, it is as if you painted a picture in my mind or made a movie step by step. I really appreciate the day to day items used to compare realization of function, reaction, and purpose. I was trying to answer a question for A & P 1’s class assignment which is “Which level of protein structure must be maintained to keep it biologically active?”
It seems one level builds upon the next supporting and protecting each level. So if one level is disrupted, the ones before and after that particular level will maintain or at least regain stability. But I didn’t get which level of the protein structure must be maintained to keep it biologically active since they each are connected.
At first after reading I thought the “Quaternary Structure” but then after further thought, wouldn’t the “Primary level” be the one to be maintained? Humm, What do you all think?
I absolutely loved and enjoyed reading your explanation of the Protein Structural Levels. The best I’ve read thus far….
Thank you for taking the time and making it comprehensible. 🙂
Thanks, this was so helpful!
Thank you so much, absolute live saver!