Introduction
Proteins (or polypeptides) are organic macromolecules consisting of amino acid subunits (monomers) that are arranged in a specific order and folded into a specific shape. These monomers of amino acids are arranged in a specific sequence known as the primary structure which is determined by the DNA of the gene coding that protein.
Amino Acids consist of a centralized C atom attached to a unique “R” functional group which dictates which of the twenty possible amino acids found in the human body it is. All amino acids contain an amino terminal and carboxyl terminal, which play a significant role its ability to form peptide bonds with adjacent amino acids.
Amino acids can form peptide bonds with neighbouring amino acids through their amino and carboxyl terminals; the condensation will remove a water molecule and construct stable polypeptide chains [1](see figure 2).
A protein’s shape and function are not only dictated by its primary structure of amino acid bonds, but also the weak intermolecular forces between the hydrogen atoms of the main amino chain the carbonyl groups (C=O); known as the secondary structure. Hydrogen bonding (an intermolecular force between H and O,N,F) results in two possible secondary structures; alpha helix or beta sheet [2].
The alpha helix, right-handed coiled or spiral conformation will be springy and flexible whereas the beta-sheet will have high tensile strength due to hydrogen bonding. These segments of the protein can further fold and supercoil into tertiary structures, which are controlled by intermolecular forces (covalent, ionic, van der Waal) between the R-groups/ side chains of the amino acids.
Proteins often have molecular weights in the thousands to 100 thousand;[3] larger proteins might be made up of several polypeptides. The interaction of two or more polypeptides to form a functional group is known as a quaternary structure; forming a detailed globular shape for a very specific activity.
Proteins have many functions throughout the body such as:
- Transport molecules (hemoglobin transports oxygen)
- Enzymatic catalysts
- Storage molecules (Ex: Iron is stored in the liver as a complex with the protein: Ferritin)
- Movement (Ex: Proteins are the major component of muscles)
- Mechanical support (Ex: Skin and bone contain collagen-a fibrous protein)
- Mediating cell responses (Ex: Rhodopsin is a protein in the eye which is used for vision)
- Antibody proteins are needed for immune protection
- Cell differentiation uses proteins (Hormones)[4]
However, proteins can be affected by certain factors such as temperature and pH, known as denaturisation. This can destroy the primary structure of the protein rendering it unable to perform its original task and proving extremely detrimental to the cell.
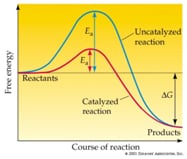
A specific type of protein, known as an enzyme is responsible for catalyzing biological reactions by a factor of 10^6. They allow complex biological reactions to happen to safe temperatures to prevent protein denaturisation and water vaporization.
It lowers the amount of energy required for the reactants to form products (ex. correct angle, sufficient force) and facilitates a successful reaction. The alternate EA transition point the enzyme provides is known as the enzyme-substrate complex.[5]
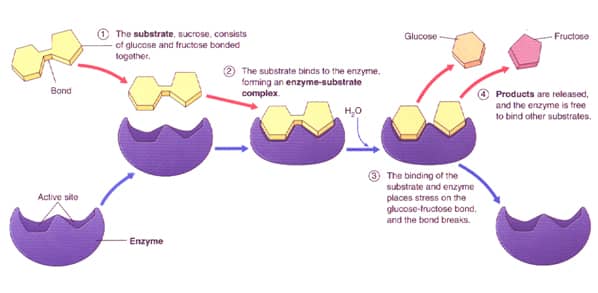
An enzyme has a specific 3-D shape which is dictated by its primary structure and how the available “R” groups interact with each other. However, the enzyme dynamically changes its shape to better accommodate the substrate which is known as the induced-fit model. The specific area in which an enzyme reacts with its designated substrate is known as the active site.
The active site for an enzyme is specific to a substrate to ensure maximum efficiency in catalyzing it toward its products; however, the enzyme is never consumed during the reaction. [6] The diagram below (figure 6) shows the steps in which the substrate binds to the active site, forms an enzyme-substrate complex, forms the product(s) and releases its.
The three-part experiment in this lab investigates the activity of two enzymes; catalase and amylase. The first and second experiments will qualitatively/ quantitatively evaluate the activity of these enzymes. Hydrogen peroxide is naturally produced in organisms as a by-product of oxygen metabolism and needs to be broken down because high levels of it are extremely toxic. [7]
H22 > O2 + H2O (catalyzed by catalase)
Amylase, a carbohydrase, catalyzes the hydrolysis of starch. The reaction can be written as: (C6H10O5)n + (n-1) H2O > nC6H12O6
The third reaction will analyze the enzyme activity rate through the decomposition of once again H22 into it products; however this time, quantitatively.
Purpose
Part 1
The purpose of this experiment is to analyze the effect of multiple variables to determine if they have an effect on the rate at which catalase catalyzes H22 decomposition.
Such factors include the re-using of the enzyme, increasing its surface area, the effect of temperature or its concentration. These sources of catalase (i.e. liver and potato) will be compared to an inorganic catalyst (mangansese dioxide) to see the effect of each variable alteration.
Part 2
The purpose of this experiment is to analyze the effect of pH on the rate in which amylase catalyzes the decomposition of starch into simpler monosaccharides. Through the use of a control and several buffers, this experiment will analyze quantitatively via Lugol Iodine indicator the effect of pH on enzyme structure and the optimum conditions for enzyme activity.
Part 3
The purpose of this experiment is to analyze the effect of substrate concentration on the activity of catalase from yeast. Through the use of control and several dilutions of H22 solutions it will quantitatively determine the correlation between [ H22] and the rate at which catalase catalyzes the substrate.
Method
Part 1
*See Guide Sheet
Part 2
*See Guide Sheet
Stagger time:
1) Devise an appropriate stagger time of about 30 seconds, so two buffers can be tested at the same time. (pH 3,5 and pH 7,9); two stop watches will be required.
2) Place the amylase into the first starch test tube and stir for several seconds; once amylase is placed into the test tube, immediately begin the stop watch.
3) At the 30 seconds mark, place the amylase into the second test tube and stir for several seconds; immediately begin the stop watch, once amylase is placed into the test tube.
4) Proceed with the experiment and dispense the solutions into their appropriate micro wells at each of their 1 minute intervals.
5) Repeat for the second pair of pH buffers being tested.
Part 3
1) Obtain all necessary materials (3% H22 solutions, H22 dilutions, stop watch, filter paper, hole punch, 50mL beaker, 25mL graduated cylinder, yeast suspension)
2) Place filter paper in yeast suspension for 2 minutes.
3) Take filter paper and place to it at the bottom of the 25mL H22 solution.
4) Observe the time it takes for the paper to rise to the top of the 25mL H22 solution.
5) Record Observations.
6) Repeat 2 more times.
7) Repeat steps 2-6 for the 1:5, 1:10, and 1:50 dilutions.
*Dilutions:
To make the dilutions for each test, a simple calculation can be performed to determine the H22 solution need:
1:5 dilution: (25mL/ 5) = 5mL of 3% H22 solution and 20mL of water
1:10 dilution: (25mL/10) = 2.5mL of 3% H22 solution and 22.5mL of water
1:50 dilution: (25mL/ 50) = 0.5mL of 3% H22 solution and 24.5mL of water
Observations
Part 1
Table 1.0: Qualitative Descriptions of the Relative Reaction Rates of Catalase
| REACTION | OBSERVATIONS | RATES OF REACTION |
| 1. Sand + H22MnO2 + H22 | No observable reactionFoaming/ hissing | –
0.5 cm rise of bubbles |
| 2. liver + H22
Potato + H22 | Vigorous reactionSlower reaction | 8.0 cm rise in bubbles5.0 cm rise of bubbles |
| 3. Used liver + fresh liverUsed liver + H22 | No observable reactionVigorous reaction |
5.0 cm rise of bubbles |
| 4. Crushed liver + H22Crushed potato + H22 | Extremely Fast | 10.0 cm rise in bubbles
8.0 cm rise in bubbles |
| 5. Boiled liver + H22Liver at 37C + H22
Liver at 0C + H22 | No observable reaction
Extremely Fast
Vigorous reaction | –
10.0 cm rise in bubbles
7.5 cm rise in bubbles |
| 6. 0.5 cm3
1.0 cm3
2.0 cm3 | FastFaster
Fastest | 4.0 cm rise in bubbles
8.0 cm rise in bubbles
14.0+ cm rise in bubbles |
Part 2
Table 2.0: Starch Test Results with Lugol’s Iodine of Amylase at various pH (3,5,7,9)
| pH | Time (min) | ||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 3 | blue | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 5 | blue | – | – | – | brown | ||||||||||
| 7 | blue | – | – | brown | |||||||||||
| 9 | blue | – | – | – | – | – | – | – | brown |
Part 3
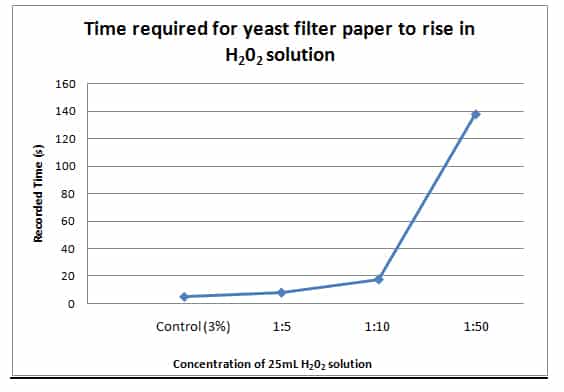
Table 3.0: Amylase Test Results with filtered paper in yeast suspension placed into H22 solutions
| Original controlled 3% of H22 solution (25mL) | 1:5 dilution of 3% H22 solution (25mL) | 1:10 dilution of 3% H22 solution (25mL) | 1:25 dilution of 3% H22 solution (25mL) | |||||||||
| Time (s) needed to rise to the top of the beaker | 1
4.98 | 2
4.87 | 3
4.65 | 1
7.41 | 2
8.01 | 3
8.65 | 1
18.34 | 2
16.98
| 3
17.55 | 1
120.0 | 2
144.04 | 3
150.18 |
| Averages (s) | 4.83 | 8.02 | 17.6 | 138.1 | ||||||||
| Control (3%) 1:5 1:10 1:50 |
Results Summary
Part 1
| REACTION | RESULTS SUMMARY |
| 1. Sand + H22MnO2 + H22 | The reaction between sand and H22 had no observable reaction, regardless of vigorous mixing and watching closely for a minute to see any significant results. The inorganic catalyst did display signs of a minimal reaction. There was hissing and foaming, but less than a centimetre (0.5 cm) of oxygen bubbles formed. |
| 2. liver + H22
Potato + H22 | Both of the sources produced vigorous and visible reactions. The liver however produced a larger column of oxygen bubbles than the potato. (Liver: 8 cm and Potato: 5 cm) (see table 1.0) |
| 3. Used liver + fresh liverUsed liver + H22 | The addition of fresh liver to used liver produced very little if no reaction at all. However the addition of more H22 solution to the used liver did produce another visible and fast reaction. (5cm of bubbles) (see table 1.0) |
| 4. Crushed liver + H22Crushed potato + H22 | Both reactions produced extremely vigorous reactions; both faster than previously observed reactions of uncrushed potato and liver. However the liver again produced a larger column of oxygen bubbles. (Liver: 10 cm and Potato: 8 cm) (see table 1.0) |
| 5. Boiled liver + H22Liver at 37C + H22
Liver at 0C + H22 | The boiled liver produced no visible reaction. There was slight discoloration of the solution and no bubbles begin formed. The liver at 0C produced a vigorous reaction producing 9cm of oxygen bubbles. The liver at 37C produced an even faster reaction and yielded an oxygen column of 12cm. (see table 1.0) |
| 6. 0.5 cm3
1.0 cm3
2.0 cm3 | Each of these produced a visible reaction which could be observed and measured. Each reaction was subsequently faster than the previous reaction and produced a larger column of oxygen bubbles. (0.5 cm3: 4 cm, 1.0 cm3= 8 cm and 2.0 cm3= +14 cm) (see table 1.0) |
Part 2
| pH | RESULTS SUMMARY |
| 1. pH= 3 | This reaction did not yield any positive results from the Lugol Iodine test. When Iodine was added to the buffered solution it turned black and remained that colour for the full duration of 15 minutes. (see table 2.0) |
| 2. pH= 5
| After 5 minutes, this reaction did produce a positive reaction to the colour light brown. (see table 2.0) |
| 3. pH= 7 | This reaction was the fastest of all 4 trials and produced a positive reaction in 4 minutes after the Lugo Iodine indicator was added. (see table 2.0) |
| 4. pH= 9 | This reaction took 9 minutes to yield results. The transition in colour between each micro well was extremely slow, but did yield a positive result to the colour light brown. (see table 2.0) |
Part 3
| [H22] | RESULTS SUMMARY |
| 1. Original 3% Solution | This was the fastest reaction to occur, taking only an average of 4.83 seconds for the filter paper disk to reach the top of the solution. (see table 3.0/ graphic 1.0) |
| 2. 1:5 Dilution
| This reaction took an average of 8.02 seconds to transpire, which was twice as long as the control test. (see table 3.0/ graphic 1.0) |
| 3. 1:10 Dilution | This reaction took an average of 17.6, about four times longer than the control test. (see table 3.0/ graphic 1.0) |
| 4. 1:50 Dilution | This reaction was by far the slowest of all 4 trials taking an average of 138.1 seconds to reach the top of the solution. (see table 3.0/ graphic 1.0) |
Error Analysis
Error: Part 1
Inaccurate liver sample sizes and finger contact
The first error that was problematic in ascertaining accurate results in part 1 was the inability to cut identical 1cm3 pieces of liver. Despite the use of a ruler, the slippery texture and irregular shape of the liver sample made it impossible to cut matching pieces, therefore certain pieces were slightly larger or smaller than others.
This proved to be a dilemma because the control variable for several parts of the experiment was altered and therefore provided inaccurate data. Having a larger piece of liver would have given the liver greater surface area in which to react with the H22 solution and therefore could have created a larger column of oxygen bubbles; having a smaller piece would have resulted in vice versa.
Also, any finger contact between the liver sample and the experimenters could have caused a transfer of oils from their fingers to the liver piece. The insoluble oils could have delayed or completely hindered the touched surfaces from catalyzing the decomposition of the H22 solution and once again provided inaccurate data.
Error: Part 2
Stagger time slightly off
This result is an entirely human error but is a problem because of the difficultly of properly managing the stagger time for this experiment. The stagger time selected by most groups was 30 seconds, giving ample time to mix the starch with amylase and then place it into its approximate micro well. However, if a group was off by 5-10 seconds, due to human inefficiency, this could have resulted in inaccurate data.
Though unlikely, being off by perhaps 10 seconds during your first stagger time would cause the timer to be off by 10 seconds each time they reach the 1 minute interval; after 6 rounds of observations the time would be off by an entire minute. This may not be significant for smaller time variations (such as 1-3 seconds) but could be greatly problematic for yielding accurate values for when the amylase is functioning properly at perhaps pH 7 or 5.
Error: Part 3
Touching filter paper
Touching the paper is once again entirely human error but could have easily transpired because the tweezers made it extremely difficult to separate the filter paper into single sheets. Touching the filter paper would have resulted in the transfer of oils from the experimenters’ hands to the filter paper and coat it with a non-water soluble layer.
This could have delayed or completely hindered the touched surfaces from catalyzing the decomposition of the H22 solution, taking more time for the filter paper to raise to the top of the solution; providing inaccurate results.
Yeast falling to bottom
This was a very simple error, which could be easily corrected. However not stirring the yeast suspension, would have resulted in the yeast particles sinking to the bottom and this could alter the concentration of the yeast on the filter paper. By not stirring it, the filter paper was submerged in a lower concentration and by stirring it; it would increase the potential concentration that could be absorbed by the filter paper.
This could create an erroneous control and provide inaccurate data. A lower concentration of the yeast would take longer to rise to the top; less oxygen bubbles would be provided from catalyzing the decomposition of the H22 solution.
Conclusion
Part 1
For the most part, the results for this experiment transpired as predicted. In the first test, with the inorganic catalyst (MnO2) and the sand, there was only a minimal reaction observed with the MnO2. The sand was inert and produced no reaction because it lacked any component to cause the H22 solution to decompose into its products; the MnO2 produced only 0.5cm (table 1) of oxygen bubbles because though it is a catalyst for this reaction, it has minimum efficiency.
In the second test, both the potato and liver reacted with visible results due to them both possessing catalase. However the reaction with the liver produced a more vigorous reaction which can be attributed to it containing more catalase than the comparative potato slice. For test three, the addition of more liver to an already catalyzed reaction of H22 yielded no results, whereas the addition of more H22 solution did.
This is because adding more catalase to the reaction but not more substrate gives the enzyme nothing to react with and therefore no reaction is produced. However the addition of more H22 solution does produce a reaction because the catalase is never consumed by the reaction it catalyzes and therefore can continue to catalyze substrate as more is added.
Crushing both the liver and potato produced a faster reaction with longer oxygen columns than the cubic pieces because this increased the surface area of the catalase. This increase in surface area allowed a greater percentage of catalase to react with the H22 solution and produce a much more vigorous result. As for the variable of temperature, boiling the liver caused it to yield no reaction because it was denatured.
The excessive heat caused the catalase’s peptide bonds to break or re-arrange, altering its active site and rendering it unable to decompose the H22 solution. The liver at 37C had an optimum reaction because this is the average temperature of our body and thus the optimum temperature for this enzyme. The liver at 0C still produced visible results; however the oxygen column they produced was smaller than the one produced when the catalase was at its optimal temperature.
Finally the increasing increments of the liver subsequently produced double the amount of oxygen bubbles for each sequential reaction. .5cm3 yielded 4cm (table 1) of bubbles, doubling to 1cm3 produced twice as many bubbles (8cm) (table 1) and 2.0cm3 produced +14cm (table 1) of bubbles. By doubling the size of liver per reaction, it doubles the amount of catalase available to react, producing twice as many oxygen bubbles rapidly.
Part 2
For this experiment, several different outcomes were recorded for the catalyzed decomposition of starch into simpler monosaccharides via amylase. At a pH of 3, there was no visible reaction observed in the micro wells, which is due to the acidity of the test tube environment. The functioning pH range for the enzyme alpha-amylase (which is found in saliva and the pancreas) is between 6.7- 7.0; with an optimum pH of 6.8 (slightly acidic).
At pH 3 (more than 1000 times more acidic) than its functioning pH range, the amylase denatured and was no longer able to bind with the substrate at its active site to catalyze the reaction. This is exactly what was observed, because during the full 15 minute observation, there was no colour change with the addition of Lugol- Iodine solution.
At the pH of 5 and 7, there were visible colour changes that transpired within the first 5 minutes (table 2). The pH 7 yielded visible results a full minute faster than the pH 5 because it was closer to the optimum pH of the enzyme. As seen in figure 14, the effectiveness of an enzyme is the greatest at its optimum pH and slowly degenerates as the surrounding pH shifts either lower or higher.
At pH 5, it was not acidic enough to completely denature the amylase structure but it did impede its ability to catalyze the reaction and therefore resulted in it taking a full minute longer to produce the light brown colour. At pH 7, it was obviously much closer to the optimum pH and therefore catalyzed the reaction at the fastest rate out of all 4 tests.
At the pH of 9 (basic) it took the amylase 9 minutes to produce the indication of starch being converted into simpler monosaccharides. It was expected that at this pH the amylase would have denatured, however a change in [OH] may have reduced the enzyme’s activity (figure 14) but it didn’t completely denature it and prevent it from catalyzing the reaction.
Part 3
During part 3, the reaction transpired in accordance with the increase or reduction of H22 concentration within the 25mL beaker. As seen in the (graphic 1), the greater the dilution of H22 solution, the more time it took for the filter paper to reach the top of the beaker. At the controlled reaction of pure 3% H22 solution it took an average of 4.83 seconds to transpire, which was the fastest of all 4 solutions.
This can be expected because the amylase located on the filter paper from the yeast suspension will be able to catalyze the greatest % concentration of H22 solution. This was made evident with the experimental data because there was more substrate present for the amylase to catalyze and therefore more oxygen bubbles produced.
As the dilution factor increased, (1:5) (1:10) (1:50) so did the amount of time it took for the filter paper to reach the top of the suspension. As seen with the experimental data from table 2 or graphic 1, each dilution, subsequently reduced the amount concentration of H22 solution per 25mL and provided less substrate in which the amylase could catalyze.
This resulted in less H22 being catalyzed to its products and less oxygen bubbles being created to raise it to the surface.
[1] Transgalactic Ltd, Initials. (2005). What is Protein?. Retrieved from http://www.bionewsonline.com/5/what_is_protein.htm
[2] Campbell, NA and Reece JB (2005) Biology. Seventh Edition. Pearson Education, Inc
[3]G., Deleage. (1994). Lecture 1: secondary structure of proteins. Retrieved from http://www.chembio.uoguelph.ca/educmat/phy456/456lec01.htm
[4] Port, T. (2003). What is an Enzyme catalyst?. Retrieved from http://biochemistry.suite101.com/article.cfm/what_is_an_enzyme
[5] Di giuiseppe , M. (2003 ). Biology 12. Toronto, Ontartio: Nelson.
[6] Di giuiseppe , M. (2003 ). Biology 12. Toronto, Ontartio: Nelson.
[7] Williams, D. (2003, July 17). The many Benefits of hydrogen peroxide. Retrieved from http://educate-yourself.org/cancer/benefitsofhydrogenperozide17jul03.shtml

this was so helpful and informative!! i obtained a result for the boiled liver, but i was’nt sure if it was correct:( this article definitely cleared my doubts! thanks again!:)