- The genetic code for a protein is defined as degenerate because there is more than 1 codon that can code for an amino acid.
- Codon bias is the probability that a given codon will be used to code for an amino acid over a synonymous codon.
- This report will be exploring codon bias for Arginine in p53’s DNA genome across human, rat, dog, and macaque
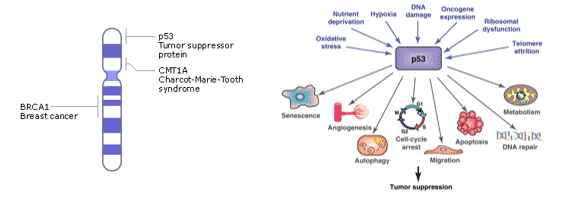
What is p53?
- p53 is a gene that codes for tumor suppression in mammalian species.
- Regulates cell division by preventing neoplasia (uncontrollable and abnormal cell growth).
- Located on chromosome 17 where it directly binds to DNA in response to stress signals.
- Structure: 393 amino acids with 4 domains that recognize damaged DNA, specific DNA sequences, and transcription factors
How Does p53 Work?
- Expression of regulator MDM2 is activated by p53. Low levels of p53 are maintained in a normal cell by the MDM2/p53 complex which triggers degradation of p53 by ubiquitin ligase.
- In the event of DNA damage, protein kinases such as ATM are activated which phosphorylate p53 at residues Ser15, Thr18, or Ser20. This will disrupt the complex and p53 levels will increase in the cell.
- After DNA is repaired, p53 will dephosphorylate and be bound to MDM2 again.
- The future to curing cancer may lie in the p53 suppressor gene. “50% of cancers have missense point mutations in the p53 gene.” (Vogelstein et al 2000). This high percentage proves that p53 is essential for survival of anti-cancerous cells.
Understanding Codon Bias
Evolutionary theories
Mutational bias versus selection
Mutational bias: codon bias exists due to non-random mutation patterns.
Selection: codon bias contributes to the efficiency and accuracy of protein expression.
Mutation Drift Model
Major codons are selected over minor codons. Minor codons play a role in genetic drift.
Effect of Codon composition on RNA/transcription/rate of translation:
- RNA secondary structure: synonymous changes of codons can alter gene expression.
- Transcription: codon optimization (matching tRNA abundances to codons).
- Rate of translation elongation: highly expressed codons translate faster than rare codons
- Effect on protein folding: the rate at which codons are translated have an impact on the protein fold
tRNA Levels and Codon Bias
commonly used codons correspond to abundant tRNA in highly expressed genes.
Hypothesis
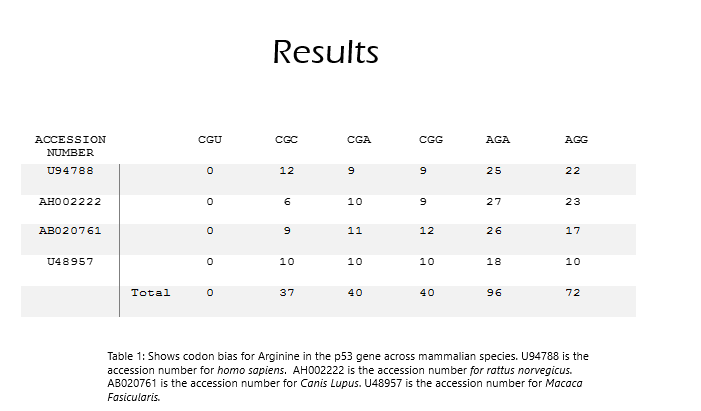
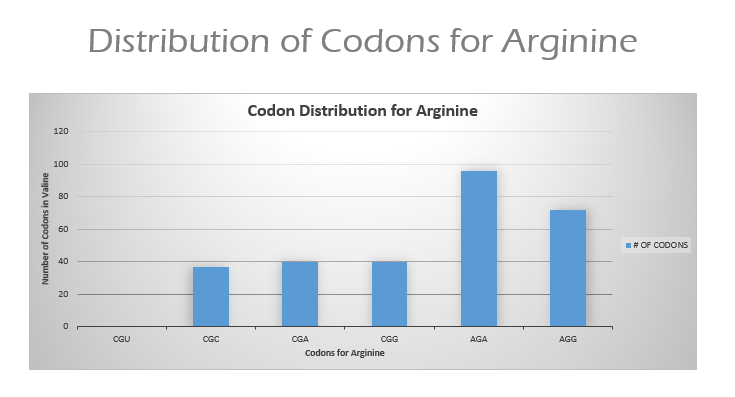
There are 6 codons that code for Arginine: CGU, CGC, CGA, CGG, AGA, and AGG in mammalian species. It is expected that these codons will be used at around the same frequency since they code for the same amino acid.
Methods
- National Center for Biotechnology (NCBI) nucleotide database was used to find the DNA genome complete CDS for humans, rat, dog, and macaque
- NCBI open reading frame finder was used to convert the DNA sequence to a polypeptide sequence.
- A chi square analysis was used to test the null hypothesis
Results
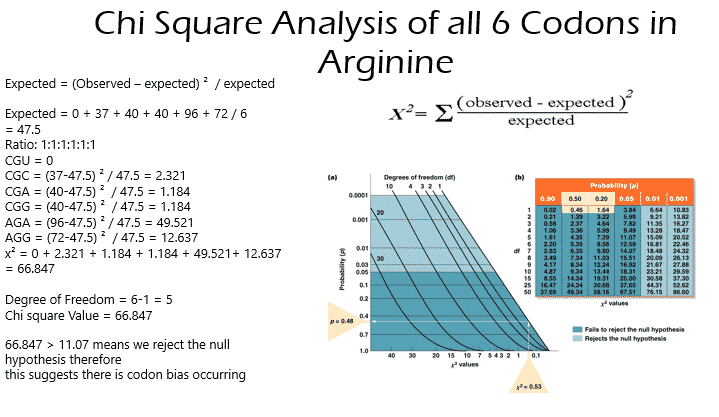
Chi Square Analysis of CGU, CGC, CGA, and CGG vs AGA and AGG
Chi squared = (Observed – expected) ² / expected
Expected = 0 + 37 + 40 + 40 /4 = 29.25
Ratio: 1:1:1:1
CGU = 0
CGC = (37-47.5) ² / 29.25= 3.770
CGA = (40-47.5) ² / 29.25 = 1.923
CGG = (40-47.5) ² / 29.25= 1.923
Degree of Freedom = 4-1 = 3
Chi square Value = 7.616
7.616 < 7.82
Fails to reject the null hypothesis
= suggests no codon bias
Chi squared = (Observed – expected) ² / expected
= 96 + 72/ 2= 84
Ratio: 1:1
AGA = (96-47.5) ² / 84 = 28.003
AGG = (72-47.5) ² / 84 = 10.360
Degree of Freedom = 2-1 = 1
Chi Square Value: 38.363
38.363 > 3.84
Rejects the null hypothesis
= suggests codon bias
Conclusion
- The 6 codons CGU, CGC, CGA, CGG, AGA, and AGG that code for Arginine rejected the null hypothesis which suggests there is codon bias amongst mammalian species.
- A chi square analysis was done on CGU, CGC, CGA and CGG and failed to reject the null hypothesis which suggests no codon bias. This was expected since the frequency at which these codons were occurring were relatively the same.
- A chi square analysis was also done on AGA and AGG which suggests codon bias.
- Researchers have different explanations for codon bias in molecular evolution. One hypothesis is that it is a genetic adaption amongst species. Others hypothesize that it may be due to a selection for alleles.
- Studies show there is a correlation between codon bias and selection in mammalian species. “Another possible selection force may be toward translation efficiency” (TE). As each codon is read by a different set of tRNAs, codons with more abundant corresponding tRNAs are likely to be translated more quickly. Therefore, synonymous and nonsynonymous codon usage affect translation elongation rate and therefore the TE of the gene” (Resso et al 2005). Selection enhances translation efficiency and accuracy in major codons. Selection also reduces translation in rare codons.
References
- Deconstructing p53 transcriptional networks in tumor suppression. Bieging, Kathryn T. et al.
- Trends in Cell Biology , Volume 22 , Issue 2 , 97 – 106. Vogelstein B, Lane D, Levine AJ. (2000). Surfing the p53 network. Nature, 408:307-310
- NCBI – National Center for Biotechnology Information … (n.d.). https://www.ncbi.nlm.nih.gov/
- Chamary J-V, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770.
- Russo A, Bazan V, Iacopetta B, Karr D, Sousi T, Gebbia N. The TP53 Colorectal Cancer International Collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol 2005;23:7518–28.